-Search query
-Search result
Showing 1 - 50 of 102 items for (author: williams & hm)
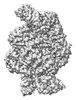
EMDB-18963: 
Structure of the SFTSV L protein in a transcription-priming state without capped RNA [TRANSCRIPTION-PRIMING (in vitro)]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Busch C, Milewski M, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M
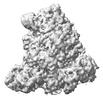
EMDB-18967: 
Structure of the SFTSV L protein in a transcription-priming state with bound capped RNA [TRANSCRIPTION-PRIMING]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Busch C, Milewski M, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-18969: 
Structure of the SFTSV L protein stalled in a transcription-specific early elongation state with bound capped RNA [TRANSCRIPTION-EARLY-ELONGATION]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Busch C, Milewski M, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-18659: 
Cryo-EM Structure of Human Kv3.1 in Complex with Modulator AUT1
Method: single particle / : Chi G, Mckinley G, Marsden B, Pike ACW, Ye M, Brooke LM, Bakshi S, Pilati N, Marasco A, Gunthorpe M, Alvaro G, Large C, Lakshminaraya B, Williams E, Sauer DB

EMDB-18660: 
Cryo-EM Structure of Human Kv3.1 in Complex with Modulator AUT5
Method: single particle / : Chi G, Mckinley G, Marsden B, Pike ACW, Ye M, Brooke LM, Bakshi S, Lakshminarayana B, Pilati N, Marasco A, Gunthorpe M, Alvaro GS, Large CH, Williams E, Sauer DB

PDB-8quc: 
Cryo-EM Structure of Human Kv3.1 in Complex with Modulator AUT1
Method: single particle / : Chi G, Mckinley G, Marsden B, Pike ACW, Ye M, Brooke LM, Bakshi S, Pilati N, Marasco A, Gunthorpe M, Alvaro G, Large C, Lakshminaraya B, Williams E, Sauer DB

PDB-8qud: 
Cryo-EM Structure of Human Kv3.1 in Complex with Modulator AUT5
Method: single particle / : Chi G, Mckinley G, Marsden B, Pike ACW, Ye M, Brooke LM, Bakshi S, Lakshminarayana B, Pilati N, Marasco A, Gunthorpe M, Alvaro GS, Large CH, Williams E, Sauer DB
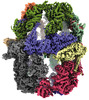
EMDB-19477: 
Saccharomyces cerevisiae FAS type I
Method: single particle / : Mann D, Grininger M, Ludig D, Sachse C

EMDB-19489: 
Tobacco mosaic virus from scanning transmission electron microscopy at CSA=2.0 mrad
Method: helical / : Mann D, Filopoulou A, Sachse C

EMDB-27820: 
Near-Atomic Resolution Structure of J-aggregated Helical Light Harvesting Nanotubes
Method: helical / : Zheng W, Deshmukh A, Caram J, Egelman EH

EMDB-27637: 
Structure of EBOV GP lacking the mucin-like domain with 2.1.1D5 scFv and 6D6 scFv bound
Method: single particle / : Yu X, Saphire EO

EMDB-27638: 
Structure of EBOV GP lacking the mucin-like domain with 9.20.1A2 Fab and 6D6 scFv bound
Method: single particle / : Yu X, Saphire EO

PDB-8dpl: 
Structure of EBOV GP lacking the mucin-like domain with 2.1.1D5 scFv and 6D6 scFv bound
Method: single particle / : Yu X, Saphire EO

PDB-8dpm: 
Structure of EBOV GP lacking the mucin-like domain with 9.20.1A2 Fab and 6D6 scFv bound
Method: single particle / : Yu X, Saphire EO

EMDB-15971: 
SARS-CoV-2 Delta-RBD complexed with Fabs BA.2-36, BA.2-23, EY6A and COVOX-45
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-27070: 
apo form Cryo-EM structure of Campylobacter jejune ketol-acid reductoisommerase crosslinked by Glutaraldehyde
Method: single particle / : Zheng S, Guddat LW

EMDB-15607: 
Structure of the SFTSV L protein bound to 5' cRNA hook [5' HOOK]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-15608: 
Structure of the SFTSV L protein stalled at early elongation [EARLY-ELONGATION]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-15610: 
Structure of the SFTSV L protein stalled at early elongation with the endonuclease domain in a raised conformation [EARLY-ELONGATION-ENDO]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-15614: 
Structure of the SFTSV L protein stalled at late elongation [LATE-ELONGATION]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-15615: 
Structure of the SFTSV L protein bound in a resting state [RESTING]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-15025: 
Leishmania tarentolae proteasome 20S subunit in complex with compound 2
Method: single particle / : Srinivas H

PDB-7zyj: 
Leishmania tarentolae proteasome 20S subunit in complex with compound 2
Method: single particle / : Srinivas H

EMDB-14885: 
OMI-42 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE GLYCOPROTEIN
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-14886: 
OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE RBD (local refinement)
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-14887: 
OMI-2 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE GLYCOPROTEIN
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-14910: 
OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13857: 
Beta049 fab in complex with SARS-CoV2 beta-Spike glycoprotein, The Beta mAb response underscores the antigenic distance to other SARS-CoV-2 variants
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13868: 
Beta-50 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13869: 
COVOX-222 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13870: 
Beta-43 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13871: 
Beta-26 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13872: 
Beta-32 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13873: 
Beta-53 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13874: 
Beta-44 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13875: 
Beta-06 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-12807: 
Apo-structure of Lassa virus L protein (well-resolved polymerase core) [APO-CORE]
Method: single particle / : Kouba T, Vogel D

EMDB-12860: 
Apo-structure of Lassa virus L protein (well-resolved endonuclease) [APO-ENDO]
Method: single particle / : Kouba T, Vogel D

EMDB-12861: 
Apo-structure of Lassa virus L protein (well-resolved alpha ribbon) [APO-RIBBON]
Method: single particle / : Kouba T, Vogel D

EMDB-12862: 
Lassa virus L protein bound to 3' promoter RNA (well-resolved polymerase core) [3END-CORE]
Method: single particle / : Kouba T, Vogel D

EMDB-12863: 
Lassa virus L protein bound to 3' promoter RNA (well-resolved endonuclease) [3END-ENDO]
Method: single particle / : Kouba T, Vogel D

EMDB-12953: 
Lassa virus L protein with endonuclease and C-terminal domains in close proximity [MID-LINK]
Method: single particle / : Kouba T, Vogel D

EMDB-12954: 
Lassa virus L protein bound to the distal promoter duplex [DISTAL-PROMOTER]
Method: single particle / : Kouba T, Vogel D

EMDB-12955: 
Lassa virus L protein in a pre-initiation conformation [PREINITIATION]
Method: single particle / : Kouba T, Vogel D

EMDB-12956: 
Lassa virus L protein in an elongation conformation [ELONGATION]
Method: single particle / : Kouba T, Vogel D

PDB-7l6o: 
Cryo-EM structure of HIV-1 Env CH848.3.D0949.10.17chim.6R.DS.SOSIP.664
Method: single particle / : Manne K, Edwards RJ, Acharya P

EMDB-23518: 
Cryo-EM map of DH851.3 bound to HIV-1 CH505 Env
Method: single particle / : Edwards RJ, Manne K, Acharya P

PDB-7lu9: 
Cryo-EM structure of DH851.3 bound to HIV-1 CH505 Env
Method: single particle / : Manne K, Edwards RJ, Acharya P

EMDB-23519: 
Cryo-EM map of DH898.1 Fab-dimer bound near the CD4 binding site of HIV-1 Env CH848 SOSIP trimer
Method: single particle / : Edwards RJ, Manne K, Acharya P

PDB-7lua: 
Cryo-EM structure of DH898.1 Fab-dimer bound near the CD4 binding site of HIV-1 Env CH848 SOSIP trimer
Method: single particle / : Manne K, Edwards RJ, Acharya P
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
